This page is will be updated with a technical summary for the sequencing efforts at Gundersen Medical Foundation's viral genome sequencing efforts for SARS-CoV-2.
For examples of media coverage of this work: News Stories
We use the Ion AmpliSeq SARS-CoV-2 Panel to sequence 237 viral specific targets encompassing the complete viral genome from SARS-COV-2-positive clinical specimens.
Current Status (updated 9/24/2020)
Viral genomes sequenced: 480
Viral genomes deposited at GISAID: 403
Viral genomes selected for sharing by public NextStrain: 49
Viral genomes available on Gundersen Nextstrain build: 514 [Contact pakenny@gundersenhealth.org for access if you are involved in COVID19 management in our region].
[Note- different quality thresholds on GISAID/NextStrain results in the number of public genomes on these platforms being lower than the total specimen number we have analyzed. Also NextStrain began downsampling strains around 4/13/20 so we anticipate that fewer of our strains will be listed there in future. We have an in-house NextStrain implementation which allows us to view and interpret the relationships between all of our sequenced specimens.]
The easiest way to begin interacting with our data is at: This Link.
Gundersen Expert Lecture Series: "Real time epidemiological SARS-CoV-2/COVID19 sequencing in the La Crosse Region" [4/15/20]:
Contact: Paraic Kenny, PhD
pakenny@gundersenhealth.org
Updates from this research study will be posted below. They represent preliminary analyses of rapidly emerging data so are subject to future revision/correction.
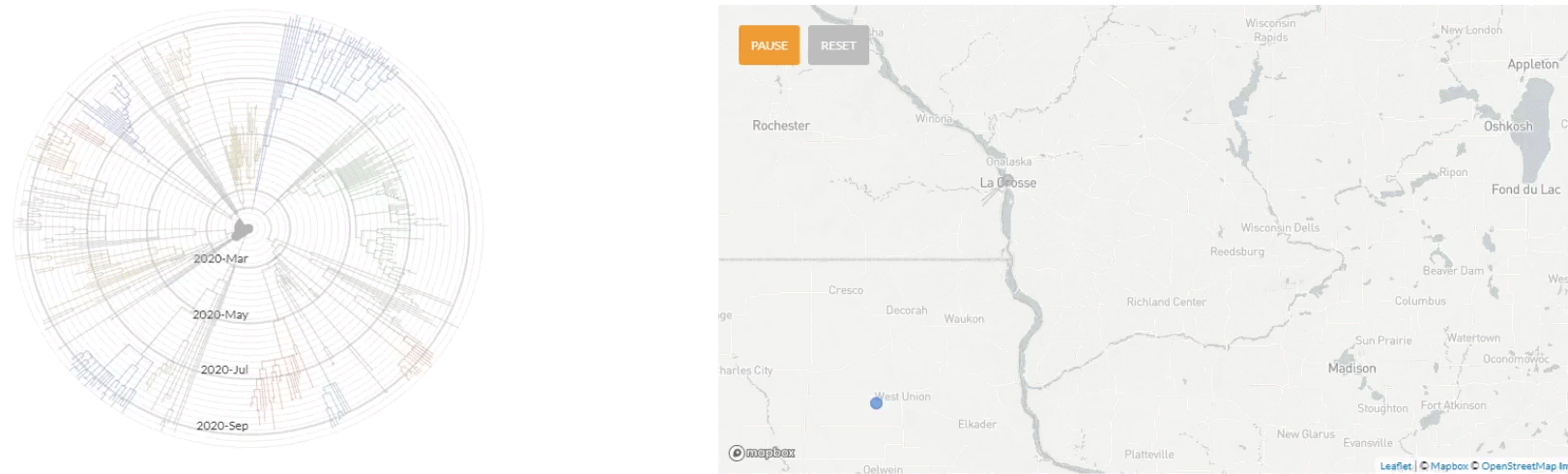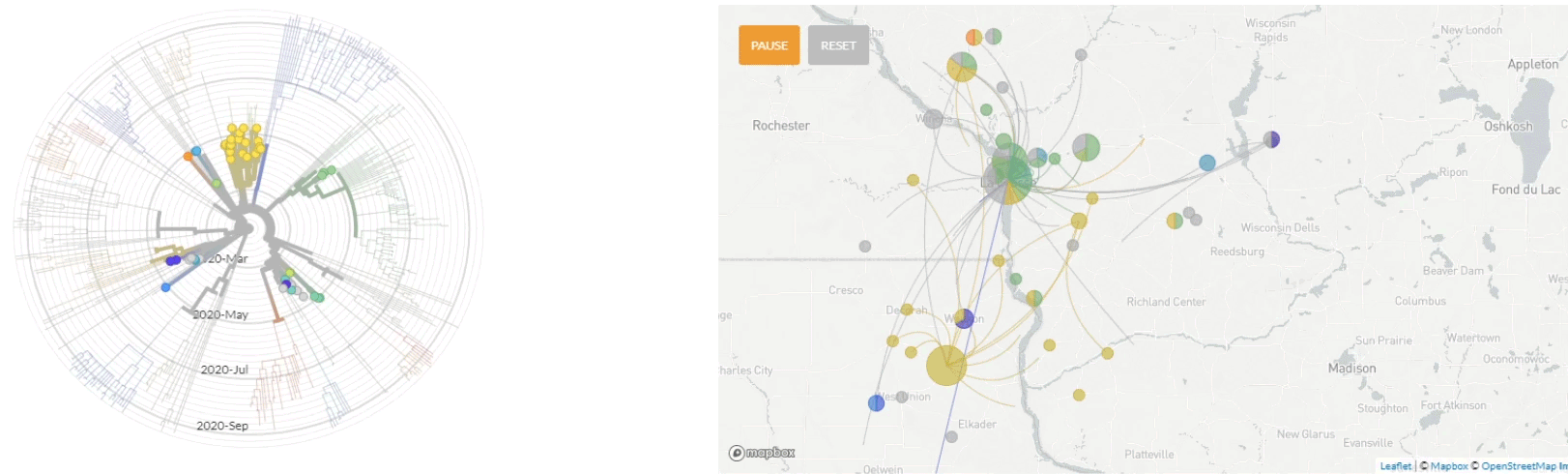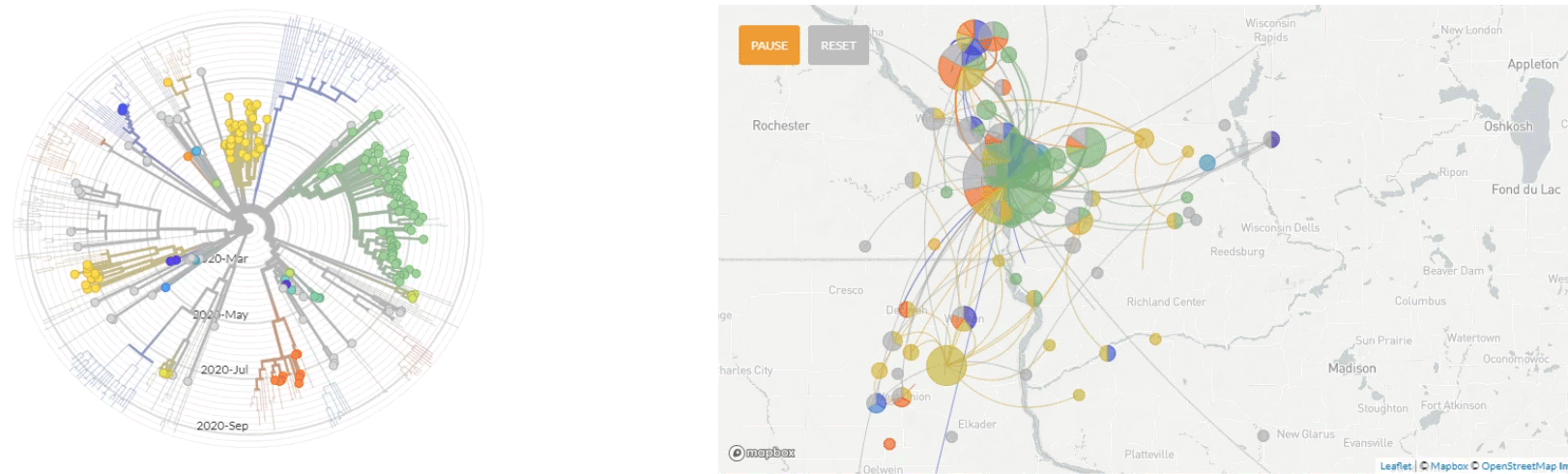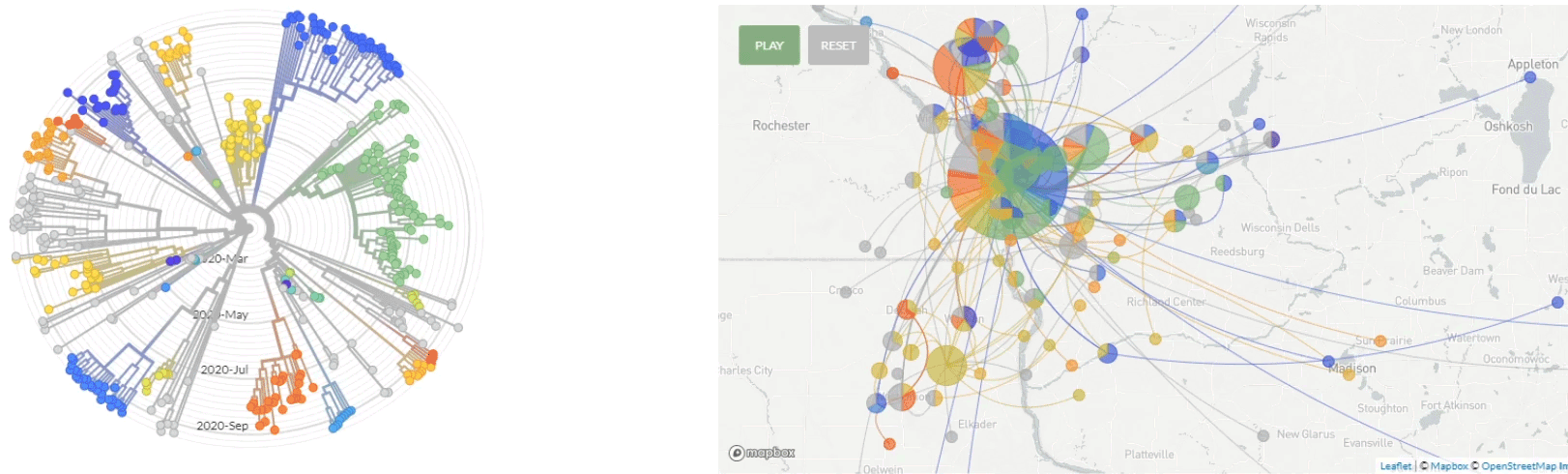
Update 10/1/2020: Use of nextstrain to map the transmission pathways within the La Crosse region (March - Sept 2020).
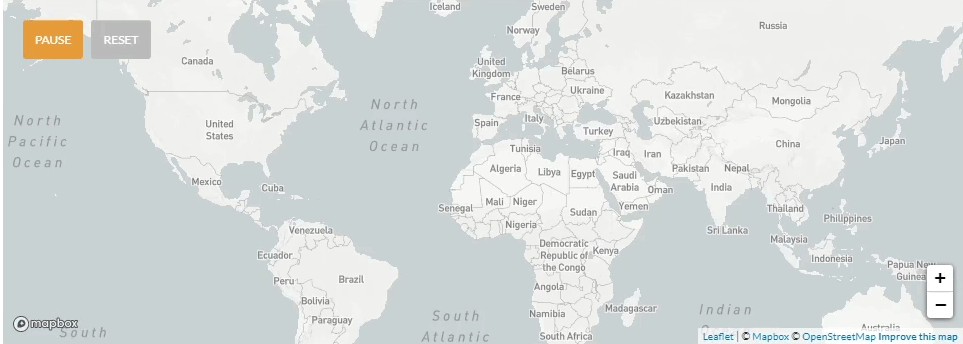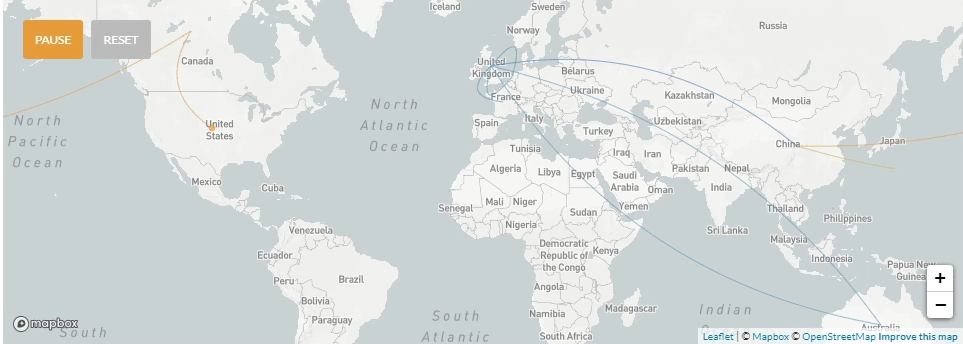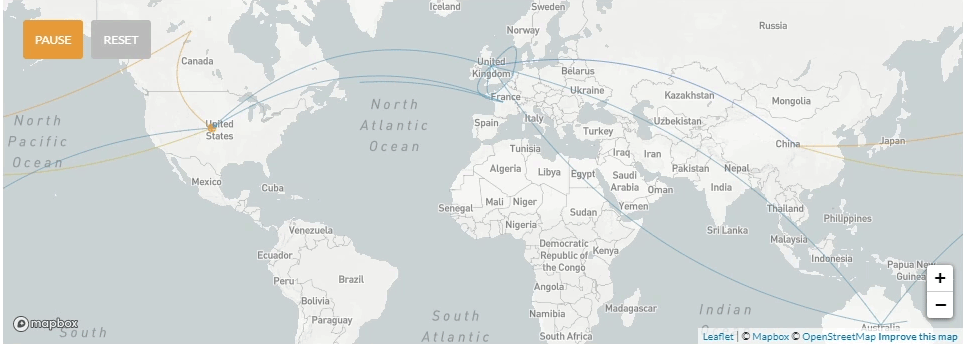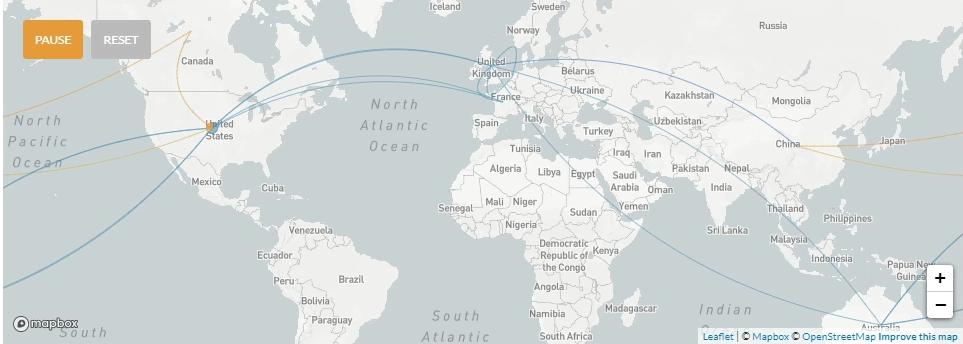
Update 4/6/2020: Use of nextstrain to map the global transmission pathways (at country-to-country resolution) taken by the viruses which caused the earliest cases in the La Crosse region.
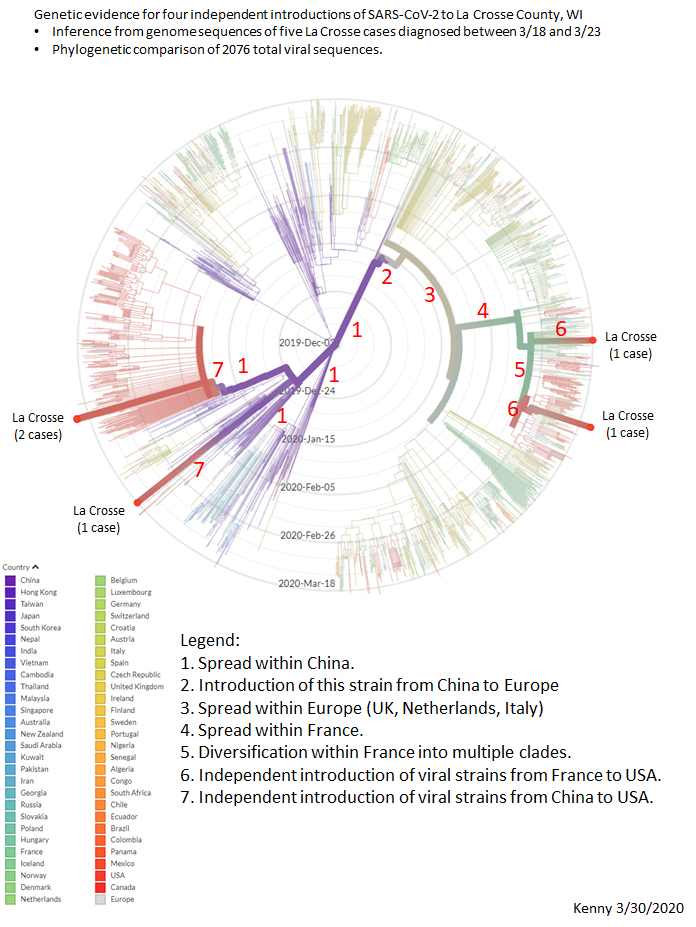
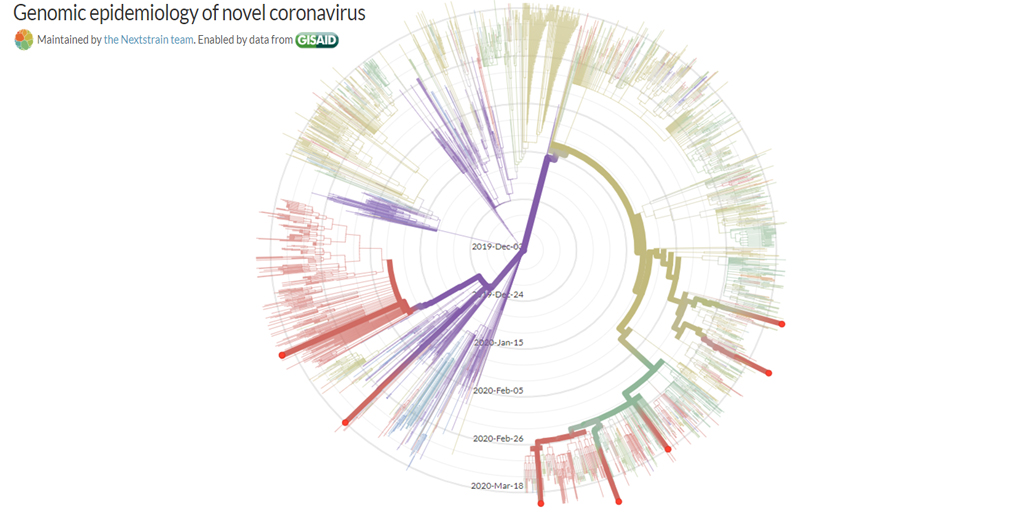
Update 4/3/2020: Phylogenetic tree including 8 early specimens from La Crosse, compared to 2894 from around the world. These data provide genetic evidence for seven independent SARS-CoV-2 introductions to La Crosse County.
Update: 3/30/20: Phylogenetic tree including 5 early viral specimens from the La Crosse region: